R Markdown Inline
The inline code is to show code verbatim, not to 'interpret' that code. What you want is completely contradictory. So you have two options: 1.
- R Markdown Code Block
- R Markdown Inline References
- R Markdown Inline Calculation
- R Markdown Inline Code
- R Markdown Inline Comments
Dynamic R code Similarly to the knitr package, you can use the markdown inline code markup or markdown code blocks to evaluate R code and insert its output into the manual page. Inline code Insert with `r `. Results appear as text without code. Built with `r getRversion`. When you render, R Markdown 1. Runs the R code, embeds.
NOTE: R Notebooks are new feature of RStudio and are only available in version 1.0 or higher of RStudio. You can download the latest version of RStudio here: https://www.rstudio.com/products/rstudio/download/.
Overview
An R Notebook is an R Markdown document with chunks that can be executed independently and interactively, with output visible immediately beneath the input.
R Notebooks are a method of literate programming that allows for direct interaction with R while producing a reproducible document with publication-quality output.
Any R Markdown document can be used as a notebook, and all R Notebooks can be rendered to other R Markdown document types. A notebook can therefore be thought of as a special execution mode for R Markdown documents. The immediacy of notebook mode makes it a good choice while authoring the R Markdown document and iterating on code; when you are ready to publish the document, you can share the notebook directly, or render it to a publication format with the Knit command.
Using Notebooks
Creating a Notebook
You can create a new notebook in RStudio with the menu command File -> New File -> R Notebook, or by using the html_notebook output type in your document’s YAML header.
By default, RStudio enables inline output (notebook mode) on all R Markdown documents, so you can interact with any R Markdown document as though it were a notebook. If you have a document with which you prefer to use the traditional console method of interaction, you can disable notebook mode by clicking the gear in the editor toolbar and choosing Chunk Output in Console.
If you prefer to use the console by default for all your R Markdown documents (restoring the behavior in previous versions of RStudio), you can make Chunk Output in Console the default: Tools -> Options -> R Markdown -> Show output inline for all R Markdown documents.
Inserting Chunks
Notebook chunks can be inserted quickly using the keyboard shortcut Ctrl + Alt + I (OS X: Cmd + Option + I), or via the Add Chunk command in the editor toolbar.
Because all of a chunk’s output appears beneath the chunk (not alongside the statement which emitted the output, as it does in rendered R Markdown output), it’s often helpful to split chunks that produce multiple outputs into two or more chunks which each produce only one output. To do this, select the code to split into a new chunk:
Then, press Ctrl + Alt + I (OS X: Cmd + Option + I):
Executing Code
Code in the notebook is executed with the same gestures you’d use to execute code in an R Markdown document:
Use the Run Chunk command, or Ctrl + Shift * Enter (OS X: Cmd + Shift + Enter) to run the current chunk.
Press Ctrl + Enter (OS X: Cmd + Enter) to run just the current statement. Running a single statement is much like running an entire chunk consisting only of that statement (see notes below on the chunk execution environment).
Use the Run All and Run Previous commands to run a batch of chunks.
The primary difference is that when executing chunks in an R Markdown document, all the code is sent to the console at once, but in a notebook, only one line at a time is sent. This allows execution to stop if a line raises an error (see notes below on handling errors).
There’s also a new Restart R and Run All Chunks command (available in the Run menu on the editor toolbar), which gives you a fresh R session prior to running all the chunks. This behavior is similar to the Knit command, which in RStudio occurs in a separate R session.
Execution Queue
When you execute code in a notebook, an indicator will appear in the gutter to show you execution progress.
Lines of code which have been sent to R are marked with dark green; lines which have not yet been sent to R are marked with light green.
If at least one chunk is waiting to be executed, you’ll see a progress meter appear in the editor’s status bar indicating the number of chunks remaining to be executed. You can click on this meter at any time to jump to the currently executing chunk.
When a chunk is waiting to execute, the Run button in its toolbar will change to a Queued icon. If you don’t want the chunk to run, you can click on the Queued icon to remove it from the execution queue.
Execution Environment
In general, when you execute code in a notebook chunk, it will do exactly the same thing as it would if that same code were typed into the console. There are however a few differences:
Output: The most obvious difference is that most forms of output produced from a notebook chunk are shown in the chunk output rather than e.g. the Viewer or Plots pane. Console output (including warnings and messages) appears both at the console and in the chunk output.
Working directory: The current working directory inside a notebook chunk is always the directory containing the notebook .Rmd file. This makes it easier to use relative paths inside notebook chunks, and also matches the behavior when knitting, making it easier to write code that works identically both interactively and in a standalone render.
You’ll get a warning if you try to change the working directory inside a notebook chunk, and the directory will revert back to the notebook’s directory once the chunk is finished executing. You can suppress this warning by using the warnings = FALSE chunk option.
If it’s necessary to execute notebook chunks in a different directory, you can change the working directory for all your chunks by using the knitr root.dir option. For instance, to execute all notebook chunks in the grandparent folder of the notebook:
This option is only effective when used inside the setup chunk. Note also that, as in knitr, the root.dir chunk option applies only to chunks; relative paths in Markdown are still relative to the notebook’s parent folder.
Warnings: Inside a notebook chunk, warnings are always displayed immediately rather than being held until the end, as in options(warn = 1).
Plots: Plots emitted from a chunk are rendered to match the width of the editor at the time the chunk was executed. The height of the plot is determined by the golden ratio. The plot’s display list is saved, too, and the plot is re-rendered to match the editor’s width when the editor is resized.
You can use the fig.width, fig.height, and fig.asp chunk options to manually specify the size of rendered plots in the notebook; you can also use knitr::opts_chunk$set(fig.width = ..., fig.height = ...) in the setup chunk to to set a default rendered size. Note however specifying a chunk size manually suppresses the generation of the display list, so plots with manually specified sizes will be resized using simple image scaling when the notebook editor is resized.
Executing Inline Chunks
An inline chunk is a chunk which is inline with the text rather than on its own line. For instance, “2 + 2 = `r 2 + 2`” will be rendered as “2 + 2 = 4”.
To execute an inline chunk in the notebook, put your cursor inside the chunk and press Ctrl + Shift + Enter (OS X: Cmd + Shift + Enter). As in the execution of ordinary chunks, the content of the chunk will be sent to the R console for evaluation. The results will appear in a small pop-up window next to the code:
In notebooks, inline R chunks can only produce text, not figures or other kinds of output. It’s also important that R code run inside inline chunks executes quickly and does not have side effects, as it’s executed whenever you save the notebook.
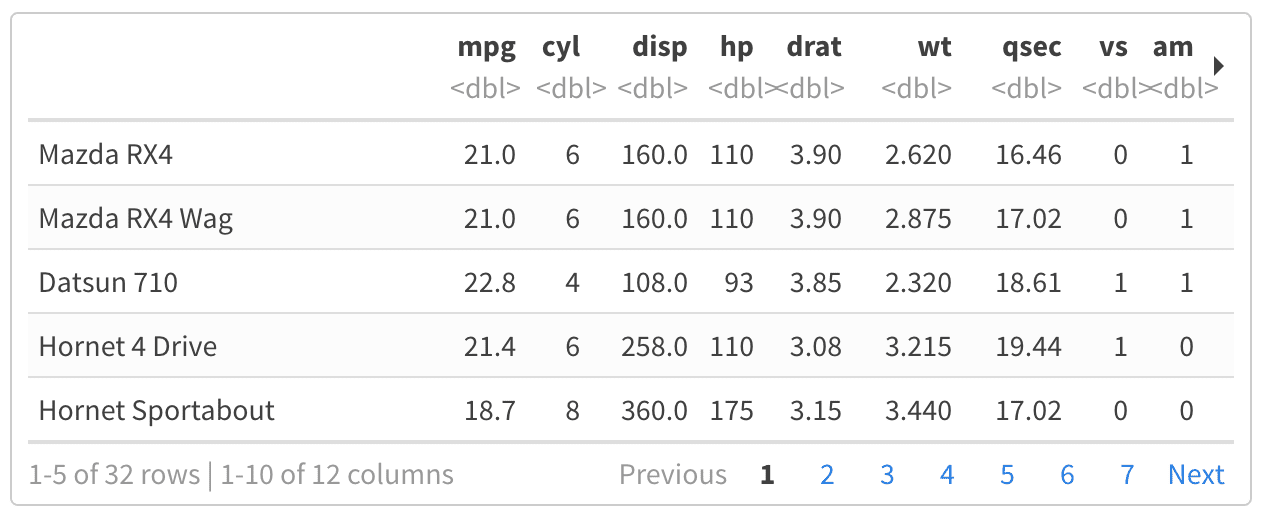
Executing External Chunks
Notebooks are typically self-contained. However, in some situations it’s preferable to re-use code from an R script as a notebook chunk, as in knitr’s code externalization. This can be done by using knitr::read_chunk in your notebook’s setup chunk, along with a special ## ---- chunkname annotation in the R file from which you intend to read code. Here is a minimal example:
example.Rmd
example.R
When you execute the empty chunk in the notebook, code from the external file will be inserted, and the results displayed inline, as though the chunk contained that code.
Chunk Output
Managing Output
When code is executed in the notebook, its output appears beneath the code chunk that produced it. You can clear an individual chunk’s output by clicking the X in the upper right corner of the output, or collapse it by clicking the chevron.
It’s also possible to clear or collapse all of the output in the document at once using the Collapse All Output and Clear All Output commands; these commands are available on the gear menu in the editor toolbar.
If you want to fully reset the state of the notebook, the command Restart R and Clear All Output will do the job.
R Markdown Code Block
See Output Storage below for more information on where chunk output is saved.

Previewing Output
Ordinary R Markdown documents are “knit”, but notebooks are “previewed”. While the notebook preview looks similar to a rendered R Markdown document, the notebook preview does not execute any of your R code chunks; it simply shows you a rendered copy of the markdown in your document along with the most recent chunk output. This preview is generated automatically whenever you save the notebook (whether you are viewing it in RStudio or not); see the section beneath on the .nb.html file for details.
When html_notebook is the topmost (default) format in your YAML header, you’ll see a Preview button in the editor toolbar. Clicking it will show you the notebook preview:
If you have configured R Markdown previewing to use the Viewer pane (as illustrated above) then the preview will be automatically updated whenever you save your notebook.
Handling Errors
When an error occurs while a notebook chunk is executing:
- Execution will stop; the remaining lines of that chunk (and any chunks that have not yet been run) will not be executed.
- The editor will scroll to the error.
- The line of code that caused the error will have a red indicator in the editor’s gutter.
If you want your notebook to keep running after an error, you can suppress the first two behaviors by specifying errors = TRUE in the chunk options.
Console Hiding
R Markdown Inline References
In most cases it shouldn’t be necessary to have the console open while using the notebook, as you can see all of the console output in the notebook itself. To preserve vertical space, the console will be automatically collapsed when you open a notebook or run a chunk in the notebook.
If you prefer not to have the console hidden when chunks are executed, uncheck Tools -> Global Options -> R Markdown -> Hide console automatically when executing notebook chunks.
Saving and Sharing
Notebook File
When a notebook .Rmd is saved, an .nb.html file is created alongside it. This file is a self-contained HTML file which contains both a rendered copy of the notebook with all current chunk outputs (suitable for display on a website) and a copy of the notebook .Rmd itself.
R Markdown Inline Calculation
You can view the .nb.html file in any ordinary web browser. It can also be opened in RStudio; when you open there (using e.g. File -> Open), RStudio will do the following:
- Extract the bundled .Rmd file and place it alongside the .nb.html file
- Open the .Rmd file in a new RStudio editor tab
- Extract the chunk outputs from the
.nb.htmlfile and place them appropriately in the editor
Note that the .nb.html file is only created for R Markdown documents which are notebooks (i.e. at least one of their output formats is html_notebook). It’s possible to have an R Markdown document which includes inline chunk output but does not produce an .nb.html file.
Output Storage
The document’s chunk outputs are also stored in an internal RStudio folder beneath the project’s .Rproj.user folder. If you work with a notebook but don’t have a project open, the outputs are stored in the RStudio state folder in your home directory (the location of this folder varies between the desktop and the server).
Version Control
R Markdown Inline Code
One of the major advantages of R Notebooks compared to other notebook systems is that they are plain-text files and therefore work well with version control. We recommend checking in both the .Rmd and .nb.html files into version control so that both your source code and output are available to collaborators. However, you can choose to include only the .Rmd file (with a .gitignore that excludes the .nb.html) if you want each collaborator to work with their own private copies of the output.
Notebook Format
While RStudio provides a set of integrated tools for authoring R Notebooks, the notebook file format itself is decoupled from RStudio. The rmarkdown package provides several functions that can be used to read and write R Notebooks outside of RStudio.
You can find documentation for these functions and additional information on the .nb.html file format in the R Notebook HTML Format article.
Learning Objectives
After completing this tutorial, you will be able to:
- Add an image to an
R markdownreport. - Describe the ideal location to store an image associated with an
R markdownreport so thatknitrcan find it when it renders a file.
What You Need
You need R and RStudio to complete this tutorial. Also you should have an earth-analytics directory set up on your computer with a /data directory with it.
R Markdown Inline Comments
Add an Image to Your Report
You can add images to an R Markdown report using markdown syntax as follows:
However, when you knit the report, R will only be able to find your image if you have placed it in the right place - RELATIVE to your .Rmd file. This is where good file management becomes extremely important.
To make this simple, let’s set up a directory named images in your earth-analytics project / working directory. If your .Rmd file is located in the root of this directory and all images that you want to include in your report are located in the images directory within the earth-analytics directory, then the path that you would use for each image would look like:
images/week3/image-name-here.png
Let’s try it with an actual image.
And here’s what that code does IF the image is in the right place:
If all of your images are in your images directory, then knitr will be able to easily find them. This also follows good file management practices because all of the images that you use in your report are contained within your project directory.
Additional Resources
